Research Profile
Research of the past decade identified a fundamental role of the intestinal microbiome in the regulation of human health. Shifts in the composition and activity of this microbial ecosystem (hereafter referred to as microbiome signatures) have been associated with a wide range of human diseases. Nevertheless, and beyond recent enthusiasm, the mechanistic implementation of microbe-host interactions into the pathogenesis of these adverse conditions is still in its infancy, and the search for disease-specific microbiome signatures with prognostic and therapeutic value using 16S rRNA gene-targeted and shotgun metagenomic sequencing is incomplete.
In this Collaborative Research Centre (CRC), we focus on the digestive tract and propose an interdisciplinary approach to characterizing the functional relevance of microbiome signatures in the context of inflammation and cancer.
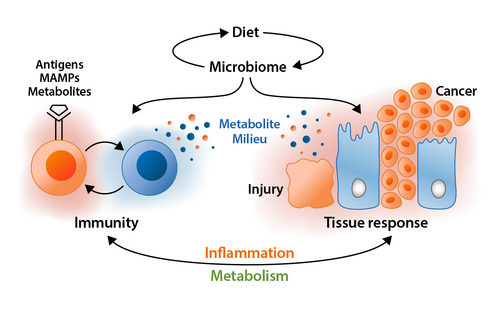
In the first funding phase we generated a unique combination of highly standardized analytical tools and novel pre-clinical models to characterize the role of microbes and their metabolites in inflammatory, metabolic and tumorigenic processes of these pathologies. In the future, we will validate the functional and clinical relevance of microbiome-related therapy in the newly established pre-clinical models (e.g. gnotobiotic mice and pigs) and defined patient cohorts. A hallmark of this research consortium is the development of synthetic bacterial consortia and the identification of clinically relevant metabolites based on complementary in silico, ex vivo and in vivo approaches. The specification of evidence-based applications for therapeutic microbiome interventions, using diet (e.g., exclusive enteral nutrition), phages and fecal microbiota transplantation will be the key objective in the upcoming years.
CRC 1371 focusses on the analytical characterization (Pillar 1) oft he human microbiome and the successive functional evaluation (Pillar 2) of therapeutic interventions, which are based on the previous findeings. In particular, we apply three comprehensive strategies to characterize the functional and clinical relevance of the intestinal microbiome in disease models and patient cohorts:
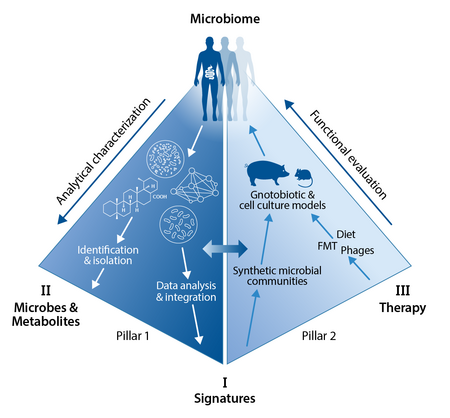
Strategy I - Characterization and functional evaluation of microbiome signatures in inflammation and cancer by applying genetically engineered and gnotobiotic animal models.
Strategy II - Identification and functional characterization of bioactive metabolites from the intestinal milieu using a comprehensive repertoire of analytical and computational methodologies.
Strategy III - Development and evaluation of microbiome-related therapeutic applications using fecal microbiota transplantation, bacteriophages and dietary interventions.