Projects
Impact of exclusive enteral nutrition on microbiome signatures and function in pediatric Crohn’s disease
Principal Investigators:
Haller, Dirk, TUM
Schwerd, Tobias, LMU
Crohn’s disease (CD) is one of the two main entities of idiopathic inflammatory bowel disease (IBD). In the second funding phase, the tandem will specify the diet-induced changes in the intestinal microbiome for therapeutic efficacy of exclusive enteral nutrition (EEN) in pediatric Crohn’s disease. EEN-associated signatures will be characterized using the combined approach of continuous culture, gnotobiotic mice and pig models. In this context, the role of dietary fiber in provoking disease relapse will be evaluated. Longitudinal bacterial- and metabolite-profiling of treatment responsive and refractory patients will be implemented to identify functionally relevant microbiome targets in response to EEN, leading to the development of protective synthetic bacterial consortia. The perspective of this project is to develop a microbiome-based maintenance therapy for pediatric CD patients.
Microbial triggers of intestinal inflammation in hosts deficient in X-linked inhibitor of apoptosis protein
Principal Investigator:
Zeissig, Sebastian, TUD
Host-microbial interactions play critical roles in the pathogenesis of inflammatory bowel disease (IBD). We have recently demonstrated that loss-of-function variants in the gene encoding X-linked inhibitor of apoptosis protein (XIAP) provide the basis for a prevalent Mendelian form of IBD. Further, we observed that mice deficient in XIAP can develop spontaneous, microbiota-induced intestinal inflammation. Here,we aim to study XIAP deficient mice as a model system to identify specific microbial triggers of intestinal inflammation in a genetically susceptible host and to delineate host pathways involved in this process.
Circadian control of microbial function in chronic intestinal inflammation
Principal Investigator:
Kiessling, Silke, TUM
Disruption of intrinsic clocks, e.g. by mismatch of the external and internal daytime in shift worker, as well as microbiota dysbiosis promote the development of gastrointestinal diseases. This project examines whether the circadian clock controls daytime-dependent fluctuations of microbiota composition and function. Microbiota transfer from will test whether the loss of microbiota fluctuations plays a key role in the development of IBD. Enhancing the clock in an IBD-relevant mouse model will determine whether the circadian clock can be a target for future strategies to treat shift-work associated syndromes.
T cell skewing in relation to intestinal dysbiosis and GvHD in allogeneic stem cell transplantation
Principal Investigators:
Busch, Dirk, TUM
Holler, Ernst, UKR
The key hypothesis of this project is that microbiota dysbiosis skews the reconstitution of a regulatory immune response in the gut and both contribute to development of graft-versus-host disease (GvHD) after allogeneic stem cell transplantation (aHSTC). This connection provides the rationale for reverting dysbiosis and the loss of regulatory T cells as a potent treatment of GvHD. We aim to identify changes in microbiota signatures, mucosal lymphocyte populations and T cell receptor repertoire composition associated with clinical outcome, taking advantage of a unique collection of gut biopsies longitudinally collected over the course of GvHD onset and development, including patients before and after fecal transplantation (FMT).
Role of innate immune signals and microbiota on intestinal epithelial regeneration in graft-versus-host disease
Principal Investigators:
Poeck, Hendrik, TUM & UKR
Ruland, Jürgen, TUM
Innate immune signaling and gut microbiota considerably affect the outcome of graft-versus-host disease (GvHD) after allogeneic hematopoietic stem cell transplantation. Using antifungal/antibacterial regimens and unique gnotobiotic animal models in an experimental setting of GvHD, we aim to understand the contribution of specific innate immune pathways and microbiota-dependent factors on intestinal stem cell function and epithelial barrier regeneration.
Mechanisms underlying intestinal microbiome orchestrated induction, maintenance and breaking of tolerance to non-self-antigens
Principal Investigators:
Biedermann, Tilo, TUM
Schnieke, Angelika, TUM
Although humans do not express α-galactose-α-1-3-galactose (α-Gal), only a few develop allergies to α-Gal+ meat. Therefore, the role of the α Gal+ intestinal microbiome for this immune tolerance in
α-Gal-/- mice and pigs will be investigated. Using gnotobiotic animals, it will be analyzed how tolerance-associated commensals modulate the immune response and allergy to α-gal. α-Gal-allergic and tolerant adults and a cohort of newborns will be investigated for α-Gal specific immune phenotypes in dependence on the intestinal microbiome composition. The long-term goal is to develop prevention or treatment strategies against food allergies by modulating the intestinal microbiome.
Microbiota-mediated modulation of adaptive immunity
Principal Investigators:
Ohnmacht, Caspar, TUM
Zehn, Dietmar, TUM
In P07, we make use of a viral infection model (LCMV) to investigate how microbial particularities of the microbiota impact the adaptive immune response to viral infections. For this purpose, we will use gnotobiotic mice colonized with natural variations of complex microbial communities and defined minimal consortia (Oligo-MM) and assess in parallel how changes in the mucosal immune system including innate immune cells and Foxp3+ regulatory T cells mechanistically impact the antigen-specific cytotoxic T cell response during acute and chronic LCMV infections.
The role of primary metabolites in the intestinal ecosystem under normal and inflamed conditions
Principal Investigators:
Jung, Kirsten, LMU
Stecher, Barbara, LMU
Inflammatory processes create metabolic and trophic niches in the gut that, in turn, affect the composition and function of the microbial ecosystem. The project P08 will investigate the role of primary luminal metabolites, specifically pyruvate, and characterize the impact of pyruvate sensing and uptake on fitness and growth of Salmonella and the individual species of the microbiota using germ-free or gnotobiotic mice colonized with the minimal bacterial consortium Oligo-MM and other mouse models of gut inflammation.
Helicobacter infection and microbiota-dependent colonic pathologies
Principal Investigators:
Deng, Li, HMGU
Gerhard, Markus, TUM
Helicobacter pylori not only induces gastric cancer but also appears to impact colonic carcinogenesis. We hypothesize that chronic H. pylori infection induces changes in the composition of the gastrointestinal microbiota, thereby promoting carcinogenesis. We will characterize the influence of
H. pylori infection on intestinal/colonic inflammation and tumorigenesis and decipher the microbiome signatures involved. These will be further explored in H. pylori-infected patients with colonic pathologies. As a therapeutic approach, we will identify specific bacteriophages using a “viral tagging” approach and test lytic phages for the treatment of H. pylori infection in mice without disruption of the endogenous microbiota.
The PIs are initiators and members of the Munich Virome Research Consortium, a new initiative for researchers working on the microbiome and virome in Munich.
Microbiota- and TLR-dependent regulation of intestinal tumor development
Principal Investigators:
Janssen, Klaus-Peter, TUM
Zeissig, Sebastian, TUD
Microbial factors can either promote or inhibit intestinal tumor development through activation of various TLR (toll-like-receptor)-dependent pathways. Here we aim to analyze whether specific microbial taxa are responsible for promoting tumor compared to tumor-suppressive effects in mouse models of colorectal cancer (CRC). We also aim to dissect the tumor-suppressive effects of TRIF/TLR3 signaling with the help of genetically engineered mouse models. The translational relevance of the preclinical findings will be evaluated using two large, independent human CRC cohorts.
Deciphering the role of tumor-site specific dysbiosis in subtypes of colorectal cancer
Principal Investigator:
Tschurtschenthaler, Markus, TUM
We aim to decipher colorectal cancer subtype-specific bacterial risk profiles and to delineate host-microbiota interactions that are relevant for tumor-initiation and progression using mouse models of BrafV637E, KrasG12D, Pik3caH1047R and Apc1638N-driven tumors. Orthotopic transplantations of organoids derived from these mice will be performed to functionally analyze the impact of various environments, i.e. SPF, germ-free and inflammatory conditions, on tumor initiation and progression with the long-term aim of targeting tumor-promoting taxa and bacterial signals.
Temporal and spatial analysis of the microbiome in a porcine model for colorectal cancer
Principal Investigators:
Saur, Dieter, TUM
Schnieke, Angelika, TUM
An existing APC1311 porcine model of colorectal cancer will be used to study causal relationships between changes in gut microbial composition and cancer progression and regression. We will experimentally manipulate the mutational load in individual polyps by in vivo and ex vivo CRISPR/Cas9 gene editing and monitor changes in microbiome composition and in lipid and bile acid metabolism during disease progression. We will also generate a new model of inflammatory bowel disease based on the TnfDARE mouse by modifying the porcine TNF gene.
Impact of dietary fat / gut microbiota interaction on intestinal lipid absorption, systemic lipid metabolism and intestinal cancer
Principal Investigators:
Ecker, Josef, UKR
Klingenspor, Martin, TUM
We aim to reveal the role of dietary fat / microbiota interaction on lipid metabolism. Mouse models with different microbiota status and intestinal cancer susceptibility will receive diets with varying dietary fat content and lipid composition. Obesity and cancer progression will be determined and bioactive lipids identified by mass-spectrometric lipid analytics, also applying stable isotope labeled tracers. Our goal is to define dietary lipid compositions and gut microbiota communities beneficial in the suppression of obesity-driven metabolic diseases and intestinal cancer.
Targeted design and manipulation of minimal bacterial consortia for microbial ecology of bile acid metabolism
Principal Investigators:
Clavel, Thomas, RWTH
Stecher, Barbara, LMU
In P14 we study the diversity of mouse intestinal bacteria involved in secondary bile acid production and investigate their impact on the host using targeted experimental models. We will establish simplified communities of cultured bacteria from the mouse gut and manipulate them in targeted manners to dissect microbe-microbe and microbe-host interactions underlying bile acid conversion both in vitro and in vivo. Thereby, we will test the causal role of secondary bile acids in gnotobiotic disease models available in this CRC.
Functional characterization of carcinogenic small molecules from the intestinal microbiota
Principal Investigators:
Gulder, Tobias, TUD
Zeller, Georg, EMBL
P16 will systematically identify biosynthetic gene clusters (BGCs) in gut microbiota through in silico mining of genomic and metagenomic resources. To infer pro-inflammatory or carcinogenic effects of the encoded metabolites, we will determine statistical enrichment in metagenomes of colorectal cancer and inflammatory bowel disease patients. Promising candidate BGCs will be cloned, heterologously expressed and characterized in cell-based assays. The impact of the identified biologically relevant metabolites on the host will be evaluated in animal models with partners of the CRC. Ultimately, this project will elucidate new links between gut microbial metabolism and host disease development
Impact of Desulfovibrio spp. and sulfur metabolism on the pathogenesis of chronic intestinal inflammation and colitis-associated cancer
Principal Investigators:
Schirmer, Melanie (TUM)
Haller, Dirk (TUM)
Persistent inflammation increases the risk of developing colorectal cancer. In this newly established project, the tandem will characterize the impact of sulfate-reducing bacteria, explicitly focusing on newly compiled human Desulfovibrio isolates, and the production of H2S on the development of chronic inflammation and colitis-associated cancer. Comparative genomic analysis of bacterial strains and the use of naturally occurring minimal consortia in germ-free mouse models provide the basis to study bacteria and host functions at the edge of inflammation and tumorigenesis. Dietary substrate- and disease-related regulation of Desulfovibrio spp. gene expression and metabolite production will be characterized in vitro and in gnotobiotic mice. Furthermore, the impact of H2S on epithelial cell functions will be determined in primary organoid cultures from mouse models and patients.
Secure integrated big data analytics
Principal Investigators:
Baumbach, Jan, UHH
Boeker, Martin, TUM
Lagkouvardos, Ilias, TUM
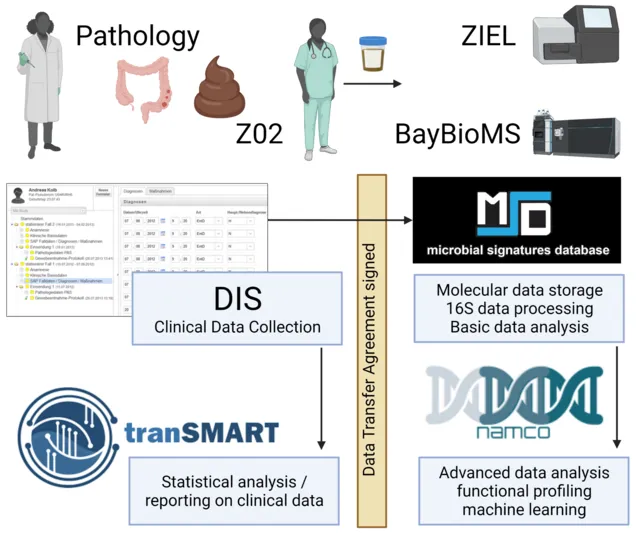
INF01 is responsible for developing infrastructure in support of (extended) clinical and research data collection, biosample management as well as translational data analytics and bioinformatics. To this end, we established an integrated platform consisting of four components (Figure)
1. DIS (Data Integration System, developed by bitcare GmbH): a secure platform supporting the collection of (extended) clinical data and patient-reported data as well as the management of well-annotated biosamples from patients relevant to this CRC, e.g. with inflammatory and neoplastic diseases.
2. MSD (Molecular Signatures Database, https://www.misigdb.org/): A microbiome signature database hosting primary human and non-human microbial metabolomic and metagenomic data, also offering analytical pipelines for processing them into signatures and profiles.
3. tranSMART: A translational data warehousing and analytics platform integrating the collected clinical and paraclinical data with associated microbiome profiles and further molecular data, e.g. RNA-seq, to support intuitive querying, data analysis and data visualisation.
4. Namco (https://exbio.wzw.tum.de/namco): A suite of CRC-specific data processing and analysis tools supporting network-enhanced pathway enrichment, biomarker discovery and systems biology-based microbiome signature mining, which will also be integrated into the translational platform.
These components interface through application programming interfaces and allow researchers and clinicians in the CRC to move from data acquisition to data analysis in a user-friendly and seamless fashion.
Clinical integration of microbiome research
Principal Investigators:
Gessner, André, UKR
Schmid, Roland M., TUM
Steiger, Katja, TUM
The Z02 clinical core unit supports integrative research into the clinical impact of microbiome signatures and provides a prospective, systematic and standardized sampling of biomaterial from patients to enable potential therapeutic options (e.g. FMT). Human material and datasets comprising all relevant information (i.e. nutritional, metabolic, microbiotic and immunologic), will be presented in a ready-to-use fashion in collaboration with INF01. We offer a comprehensive multiscale collection of patient data combined with digitalized histopathology, microbiome analyses of stool and tissue samples and, as well as optionally, human tissue sequencing data in a pseudonymized, research-applicable manner.
Integrated Research Training Group
Principal Investigator:
Klingenspor, Martin, TUM
The Integrated Research Training Group (IRTG) will provide PhD students and postdocs with a project-related qualification program dedicated to the integration of knowledge and methodologies from multiple disciplines. It will offer training opportunities to enable cross-disciplinary approaches and promote rapid progress of the individual research projects. The IRTG will foster interaction and collaboration of CRC teams at local or distant sites. Our diverse agenda of excellent academic and hands-on training will promote self-reliance in experimental and clinical research, originality in thinking, and innovation in research.
Central tasks of the Collaborative Research Centre
Principal Investigator:
Haller, Dirk, TUM
Central tasks of the CRC 1371 are coordinated and supervised by Dirk Haller in agreement with the CRC 1371 executive board. Tasks in Z04 include administrative coordination of the scientific consortia and IRTG, including the organization of CRC 1371 office, retreats, symposia, summer school, and guest lectures as well as all public relations and dissemination activities. An important task is ensuring gender equality, family and early career support. The CRC 1371 office closely interacts with the CRC 1371-specific Gender Equality Team. Finally, Z04 coordinates scientific services for microbiome and metabolite profiling.